Accurate proteome-wide missense variant effect prediction with AlphaMissense
Por um escritor misterioso
Descrição

PDF] Improved pathogenicity prediction for rare human missense variants

Predicting the pathogenicity of missense variants using features derived from AlphaFold2

CIMB, Free Full-Text

Google DeepMind Says Its New Artificial Intelligence Tool Can Predict Which Genetic Variants Are Likely to Cause Disease

Accurate prediction of protein tertiary structural changes induced by single-site mutations with equivariant graph neural networks

Predicting functional effect of missense variants using graph attention neural networks
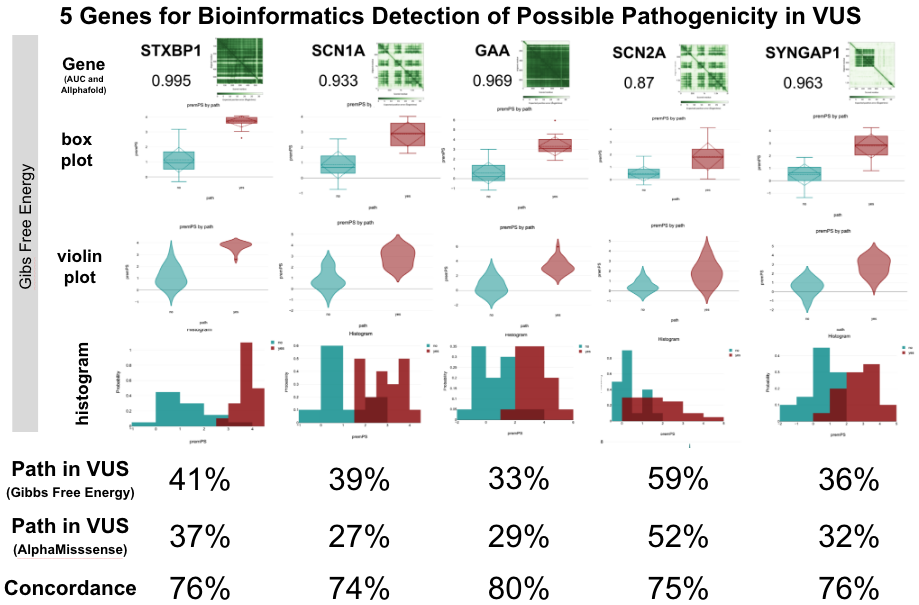
How Gene Data Sheds Light on Epilipesy

Missense Variants Reveal Functional Insights Into the Human ARID Family of Gene Regulators - ScienceDirect
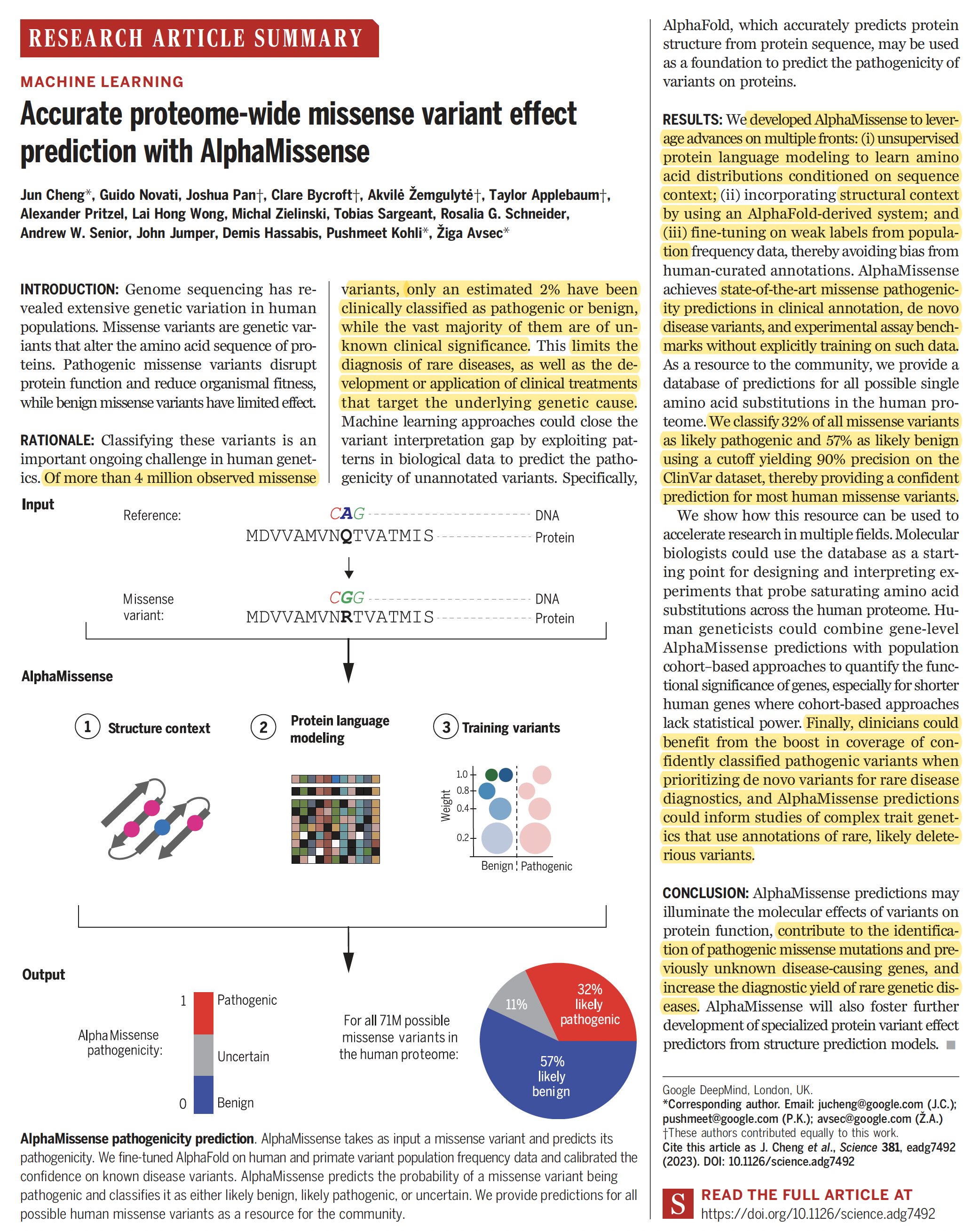
Eric Topol on X: Just out @ScienceMagazine #AlphaMissense—building on #AlphaFold—based on unsupervised learning #AI, predicts impact of all 71 million human missense variants for disease-causing potential, across entire human proteome; open-source
de
por adulto (o preço varia de acordo com o tamanho do grupo)







